Numerous methods for amplifying nucleic acids were created in the 1980s, and since then, these methods have been enhanced, and additional methods have been developed for amplification. These include transcription-mediated amplification (TMA), ligase chain reaction (LCR), nucleic acid sequence-based amplification (NASBA), strand displacement amplification (SDA), and linear linked amplification. Despite their development and refinement, none of these techniques have gained the same broad research applications as PCR and this may be due to the simplicity and affordability of PCR.
PCR is often referred to as a DNA photocopier, and while it may seem simple, it is, in fact, a complicated process that involves many reactants. Some of these reactants, such as dNTPs and primers, maintain a stable concentration throughout the reaction, while others, like DNA polymerase, may become limiting. Temperature and pH changes can cause significant fluctuations in the dynamics of various molecular interactions.
Despite its complexity, PCR is an incredibly powerful and versatile tool for DNA manipulation and analysis. Since its creation, it has transformed the field of molecular biology and enabled the isolation of genes from any organism. PCR is now an essential tool for genome sequencing projects and is used for determining DNA sequence data as well as the study of genes. Many scientific papers describe novel applications or techniques of PCR for use in research and PCR-based diagnostics.
Listed are some of the common types of nucleic acid amplification techniques.
Loop-Mediated Isothermal Amplification (LAMP):
LAMP is a novel method for amplifying genes that offer rapidity, simplicity, and high specificity. Several tests based on this method have been developed that maintain simplicity throughout all steps, from nucleic acid extraction to amplification detection. The LAMP reaction involves the amplification of samples at a constant temperature through two types of elongation reactions that occur at the loop regions. The LAMP reaction has a broad range of potential applications, including point-of-care testing, genetic testing in resource-limited settings, and rapid testing of food products. The sequential progression of the LAMP amplification reaction is rapid, contributing to its high amplification efficiency. Furthermore, the LAMP method can amplify small quantities of a gene in a short period.
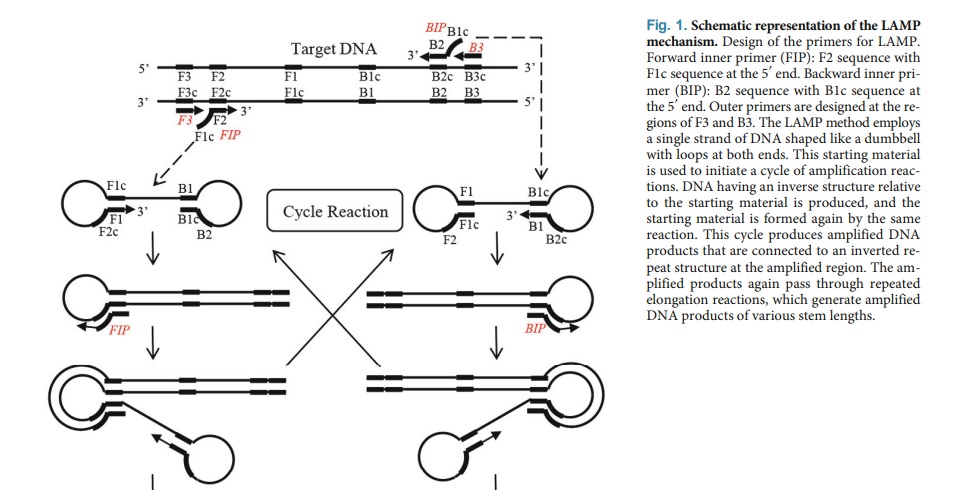
The primary distinctions between LAMP and PCR are:
- Reaction conditions: LAMP amplifies DNA under isothermal conditions while PCR requires a thermal cycler to cycle through different temperatures.
- Primers: LAMP uses four or six primers, while PCR uses two primers.
- Amplification: LAMP amplifies the target DNA rapidly and efficiently, while PCR has some limitations in amplifying certain DNA sequences.
- Detection: LAMP products can be detected visually by turbidity, colorimetry, or fluorescent dyes, while PCR products are typically detected using gel electrophoresis or by hybridization with probes.
- Cost: LAMP is relatively inexpensive compared to PCR since it does not require a thermal cycler or sophisticated equipment.
Nucleic Acid Sequence-based Amplification (NASBA):
NASBA is an isothermal amplification technique that enables the rapid detection and amplification of RNA sequences in a sample. The NASBA involves the conversion of RNA to cDNA by reverse transcriptase, followed by the synthesis of RNA from the cDNA template by RNA polymerase. The newly synthesized RNA can then be used as a template for further amplification in subsequent NASBA cycles.
NASBA is not commonly used because the commercial kits are expensive, and there are challenges in creating a dependable NASBA master mix in-house. So far, the most common use of NASBA has been for virus detection and less frequently for the detection of bacterial pathogens.
The main differences between NASBA and PCR are:
- Reaction conditions: NASBA amplifies RNA under isothermal conditions (i.e., constant temperature), while PCR requires a thermal cycler to cycle through different temperatures.
- Primers: NASBA uses two primers and one probe, while PCR uses two primers.
- Amplification: NASBA can amplify RNA rapidly and efficiently, while PCR has some limitations in amplifying certain RNA sequences.
- Detection: NASBA products can be detected visually by turbidity, colorimetry, or fluorescent dyes, while PCR products are typically detected using gel electrophoresis or by hybridization with probes.
- Sensitivity: NASBA is more sensitive than PCR due to the high amplification efficiency and use of multiple primers.
Transcription-mediated Amplification (TMA):
The TMA process involves three main steps: target capture, transcription, and amplification. In the first step, the target RNA or DNA is captured by a specific capture probe that is complementary to the target sequence. In the second step, the captured target is converted into multiple copies of RNA by a reverse transcriptase enzyme. The reverse transcriptase synthesizes a complementary DNA (cDNA) strand using the captured target as a template. The cDNA strand is then used as a template for RNA synthesis by RNA polymerase enzyme. In the final step, the amplified RNA is detected using a labeled probe that is specific to the RNA product.
TMA has several advantages over other nucleic acid amplification techniques. It is highly sensitive, and capable of detecting low levels of target RNA or DNA. TMA is also rapid, with results available in less than two hours.
TMA has been used for the diagnosis of infectious diseases such as HIV, hepatitis C, and Zikavirus, as well as for the detection of genetic mutations associated with cancer and genetic disorders.
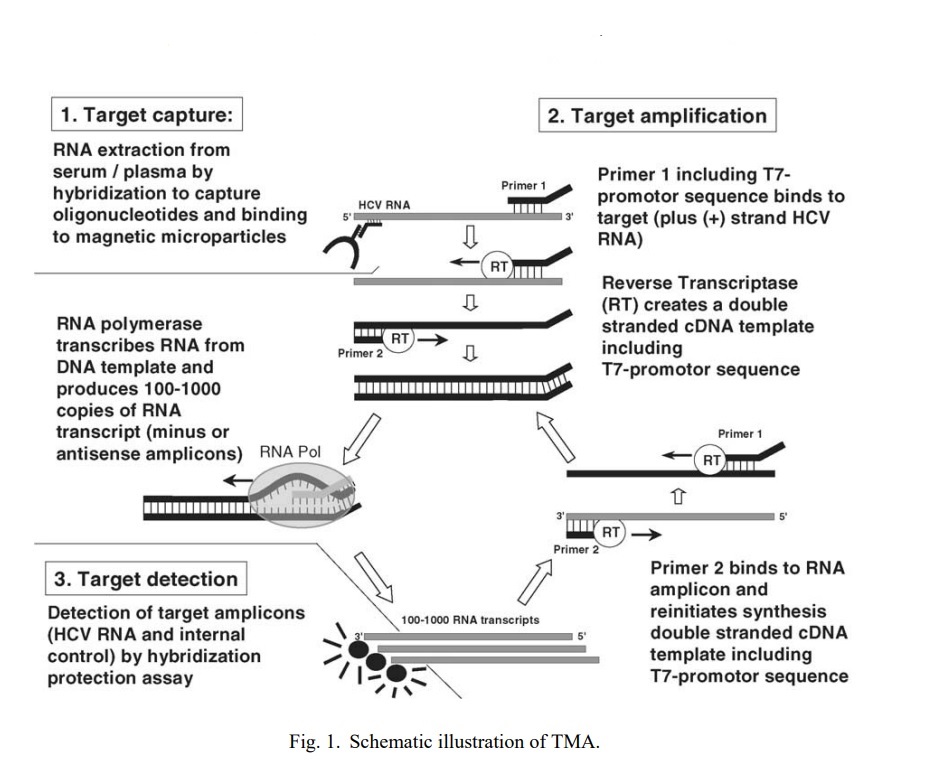
Some of the distinctions between TMA and PCR are:
- Amplification method: TMA amplifies RNA via a transcription step followed by amplification using reverse transcriptase and RNA polymerase, while PCR amplifies DNA using DNA polymerase.
- Reaction conditions: TMA amplifies RNA under isothermal conditions (i.e., constant temperature), while PCR requires a thermal cycler to cycle through different temperatures.
- Detection: TMA products can be detected visually by turbidity, colorimetry, or fluorescent dyes, while PCR products are typically detected using gel electrophoresis or by hybridization with probes.
- Sensitivity: TMA is more sensitive than PCR due to the high amplification efficiency and use of multiple primers and probes.
- Specificity: TMA is highly specific to the target RNA sequence, with a reduced risk of nonspecific amplification compared to PCR.
Reverse Transcription Polymerase Chain Reaction (RT-PCR):
RT-PCR is a highly sensitive and specific technique that can detect very small amounts of RNA in a sample, making it a valuable tool for many molecular biology applications. It is based on the principle of converting RNA molecules into complementary DNA (cDNA) molecules using the enzyme reverse transcriptase. The approach includes utilizing a primer that is attached to the RNA of significance. In the case of mRNA, the primer typically takes the form of a synthetic oligo, a combination of random hexamers, or a synthetic DNA oligonucleotide that matches a particular transcript. This blend of DNA and RNA functions as a pattern during the reverse transcription process, in which the enzyme reverse transcriptase (RT) generates a single-stranded copy of a section of the target RNA molecule called cDNA.
RT-PCR is commonly used to detect the presence of various types of RNA, including messenger RNA (mRNA), precursor mRNA (pre-mRNA), and non-coding RNA.
Few fundamental distinctions between RT-PCR and PCR are:
- Purpose: PCR is used to amplify DNA sequences, while RT-PCR is used to amplify RNA sequences.
- Reverse transcription: RT-PCR involves an additional step called reverse transcription, where RNA is converted to complementary DNA (cDNA) using reverse transcriptase enzyme before the PCR amplification step. PCR does not involve this step.
- Sensitivity: RT-PCR is more sensitive than PCR since it can detect even small amounts of RNA. In contrast, PCR is less sensitive since it detects DNA only.
- Application: RT-PCR is commonly used in applications such as gene expression analysis, viral detection, and quantification of RNA transcripts. PCR, on the other hand, is used for gene cloning, DNA sequencing, and genotyping.
Strand Displacement Amplification (SDA):
SDA is a DNA amplification technique that uses a combination of DNA polymerase and restriction endonuclease to selectively amplify specific DNA sequences. The technique was first described in 1992 and has since been widely used in research and diagnostic applications.
The SDA reaction begins with the binding of primers to the target DNA sequence, followed by the action of a restriction endonuclease to cleave the DNA. The DNA polymerase then extends the primers and displaces the single-stranded DNA produced by the restriction enzyme cleavage. This displaced strand becomes a template for another cycle of amplification, resulting in exponential amplification of the target DNA sequence.
However, some key differences between SDA and PCR are:
- Target sequence: SDA can amplify both single-stranded DNA (ssDNA) and double-stranded DNA (dsDNA), while PCR amplifies only double-stranded DNA.
- Amplification mechanism: SDA uses a combination of enzymes (such as polymerases and exonucleases) and single-stranded binding proteins to amplify DNA, whereas PCR uses a thermostable DNA polymerase enzyme and a set of primers.
- Sensitivity: SDA is more sensitive than PCR since it can detect even a single copy of the target DNA sequence. In contrast, PCR has a lower sensitivity and requires at least several copies of the target sequence.
- Product size: SDA typically generates shorter amplified DNA products (less than 1 kb) compared to PCR.
Multiple Displacement Amplification (MDA):
MDA is a commonly used method for whole genome amplification (WGA) and single-cell whole genome (scWGA) analysis. The technique involves the binding of random hexamers to denatured DNA, which is then subjected to strand displacement synthesis at a constant temperature. Unlike other DNA polymerases that require higher reaction temperatures, MDA uses shorter primers that can be hybridized to the template at an optimal temperature of about 30 °C. The amplified DNA produced by MDA has a higher molecular weight and better genome coverage. To further enhance the performance of MDA, scientists have developed customized approaches such as microwell MDA (MIDAS), emulsion MDA (eMDA), and microchannel MDA (μcMDA).
Listed below are some of the primary differences between MDA and PCR:
- Amplification mechanism: MDA uses a DNA polymerase with high processivity and strand-displacing activity, along with random primers, to amplify DNA. In contrast, PCR uses a thermostable DNA polymerase enzyme and a set of primers.
- Sensitivity: MDA is more sensitive than PCR since it can detect even a single copy of the target DNA sequence. In contrast, PCR has a lower sensitivity and requires at least several copies of the target sequence.
- Bias: MDA can introduce amplification bias due to random priming, leading to the over-representation of certain DNA sequences. PCR can also introduce bias due to primer specificity, but this can be minimized through careful primer design.
Conclusion
Nucleic acid amplification techniques have revolutionized many fields of research and have led to advances in medical diagnostics and personalized medicine.
While PCR is still the most commonly utilized method for amplifying nucleic acids, there are several other technologies available for amplification and detection. The selection of an appropriate method will depend on the sample’s characteristics and the biological query being studied. Additionally, when choosing a nucleic acid amplification technique for a particular application, factors such as sensitivity, specificity, cost, and ease of use should also be taken into account.
References
- PCR by Mike McPherson, Simon Møller
- Fakruddin M, Mannan KS, Chowdhury A, Mazumdar RM, Hossain MN, Islam S, Chowdhury MA. Nucleic acid amplification: Alternative methods of polymerase chain reaction. J Pharm Bioallied Sci. 2013 Oct;5(4):245-52. doi: 10.4103/0975-7406.120066. PMID: 24302831; PMCID: PMC3831736.
- Monis, P. T., & Giglio, S. (2006). Nucleic acid amplification-based techniques for pathogen detection and identification. Infection, Genetics and Evolution, 6(1), 2–12. doi:10.1016/j.meegid.2005.08.004
- Na Lu, Yi Qiao, Zuhong Lu, Jing Tu, Chimera: The spoiler in multiple displacement amplification, Computational and Structural Biotechnology Journal ISSN 2001-0370,
- https://cshprotocols.cshlp.org/content/2014/11/pdb.prot080887.short
- Notomi, T., Mori, Y., Tomita, N., & Kanda, H. (2015). Loop-mediated isothermal amplification (LAMP): principle, features, and future prospects. Journal of Microbiology, 53(1), 1–5. doi:10.1007/s12275-015-4656-9