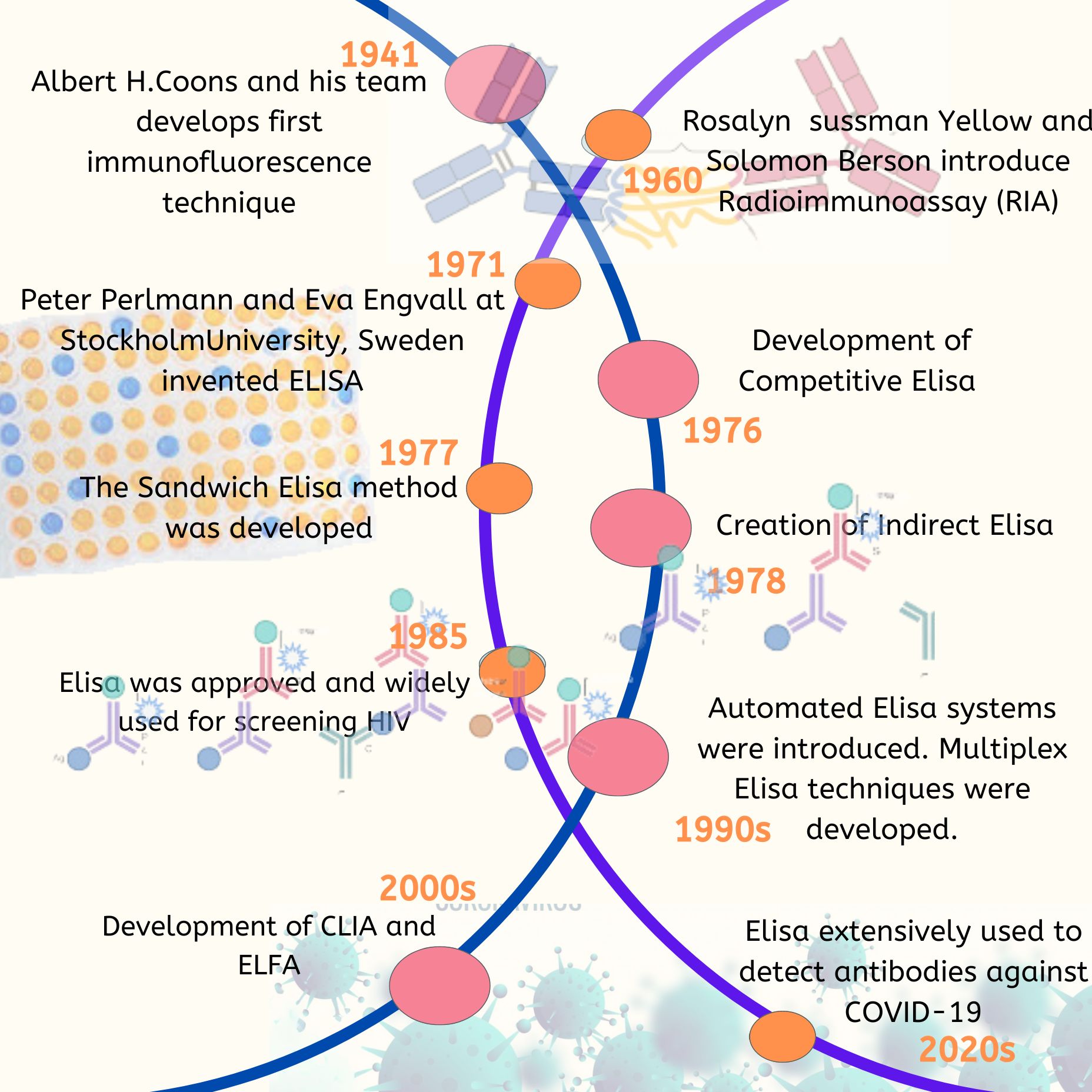
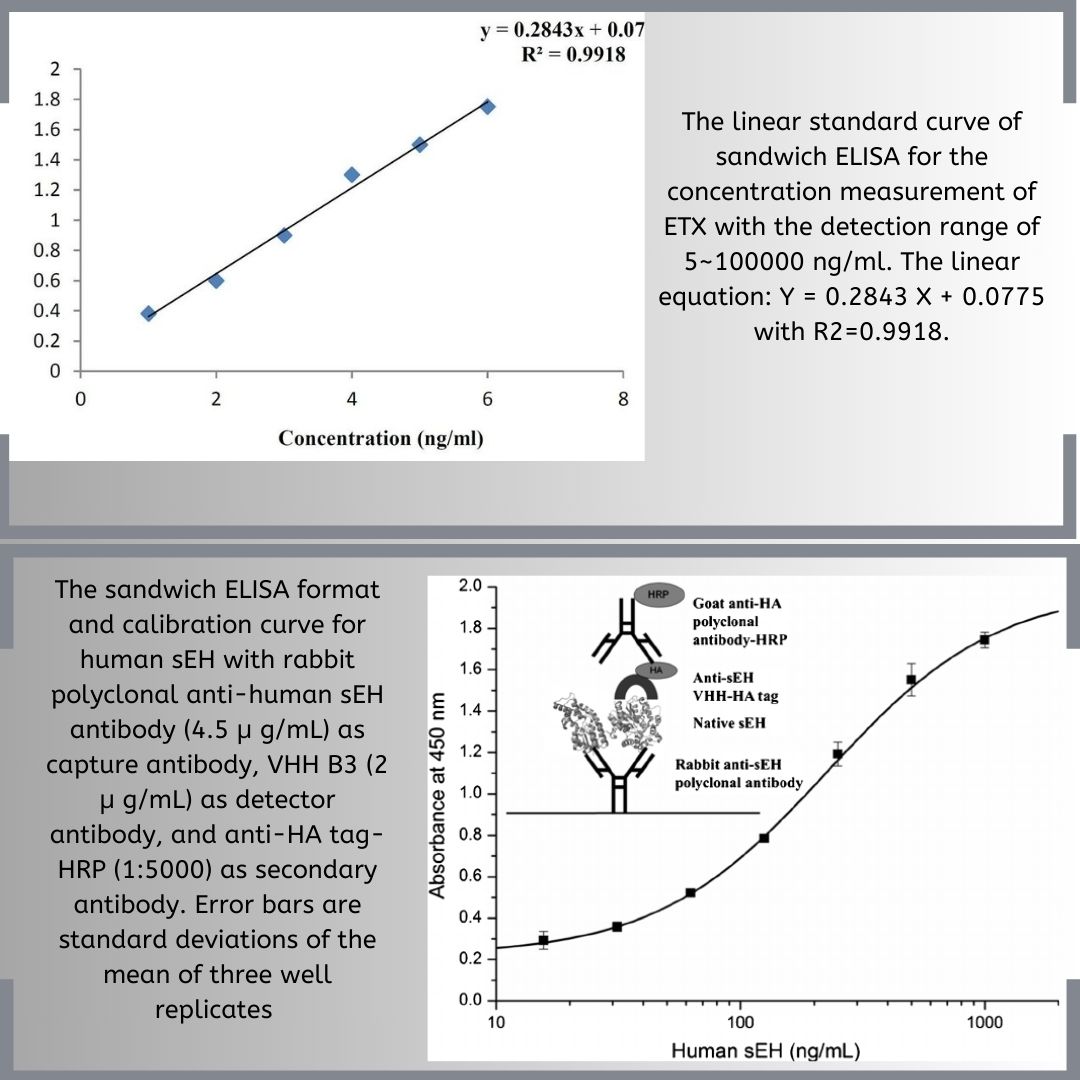
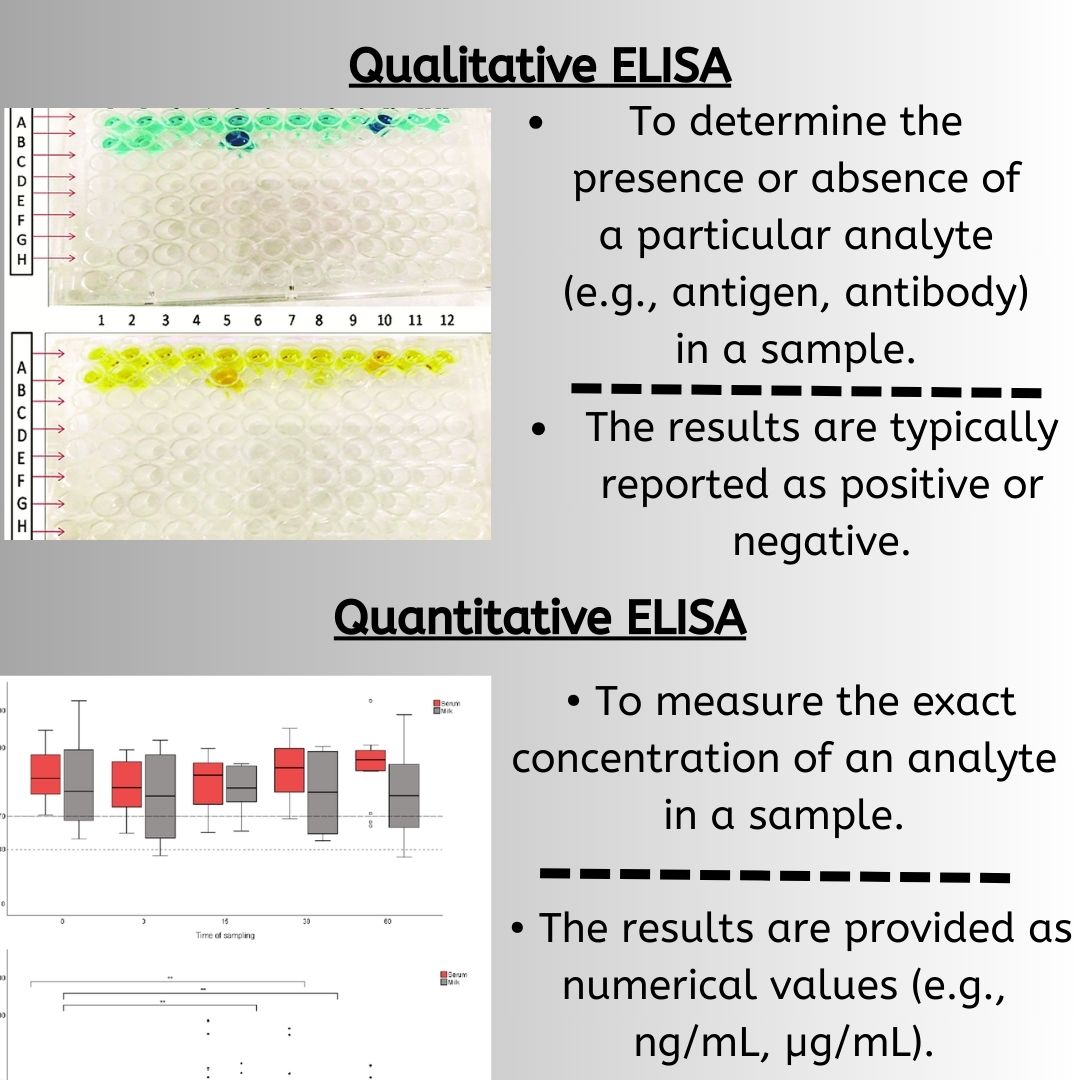
Sensitivity reflects a test’s ability to correctly identify individuals with a disease or condition, while specificity measures its ability to correctly identify those without the condition, crucial for minimizing false positives and false negatives. These factors are vital for diagnostic accuracy and performance. The method used, along with the concentration of antigens and antibodies, plays a key role in achieving the desired sensitivity and specificity. Proper preparation of samples, including serum samples, and purification of proteins can significantly impact these metrics.
Sensitivity reflects a test’s ability to correctly identify individuals with a disease or condition, while specificity measures its ability to correctly identify those without the condition, crucial for minimizing false positives and false negatives. These factors are vital for diagnostic accuracy and performance. The method used, along with the concentration of antigens and antibodies, plays a key role in achieving the desired sensitivity and specificity. Proper preparation of samples, including serum samples, and purification of proteins can significantly impact these metrics.
Numerically, sensitivity is calculated as the proportion of true positive results among all individuals with the condition, while specificity measures true negatives among those without the condition, often expressed as percentages. False positive and false negative rates are important considerations in the analytical evaluation of any assay. The threshold or cutoff point used in the assay directly affects these values, with variations observed depending on the concentration and parameters tested. Sensitivity in ELISAs varies by type (competitive, indirect, sandwich), antigens, and mAbs used, requiring experimental determination for proper validation. The information gained from these assays is critical for assessing diagnostic reliability, especially when dealing with varying patient age groups and disease symptoms. Analysis of these parameters helps in the accurate identification and measurement of biomarkers.
Competitive ELISAs are effective for low molecular weight antigens (<10,000 Daltons), including small molecules, peptides, and steroids, offering sensitivity for picomolar analytes like cAMP in cell lysate. The concentration of these analytes can vary significantly between studies and regions. ELISA assay specificity hinges on validated antibodies that prevent cross-reactivity, crucial for reliable antigen detection across diverse sample types, such as cells and tissue extracts. These assays must be compared against a reference or gold standard to ensure they meet the necessary standards of diagnostic performance, often summarized in a table or figure for clear presentation.

|
High-Affinity Antibodies: |
|
|
Low-Affinity Antigens: |
|
|
Signal Boosting: |
|
|
New Signal Systems: |
|
|
More HRP in Kits: |
|
|
Estimating Detection Limits: |
|
|
Microfluidic ELISA: |
|
|
Digital Immunoassays: |
|
|
Recombinant B-Cell Multi-Epitopes: |
|
|
Customized ELISA in Racing Labs: |
|
|
Nucleic Acid Amplification Assays: |
|
|
Aptamer Specificity in ELISA: |
|
|
Advanced Antibody Strategies in Proteomics: |
|
Explore Our Full Range of ELISA Kits
Whether you’re testing human, animal, or plant samples, MyBioSource offers over 1 million ELISA kits covering thousands of analytes across every major species.
References
- Xiong, Y., Leng, Y., Li, X., Huang, X., & Xiong, Y. (2020). Emerging strategies to enhance the sensitivity of competitive ELISA for detection of chemical contaminants in food samples. TrAC Trends in Analytical Chemistry, 126, 115861.
- Li, H., Li, Y., & Xia, Y. (2021). Advance of modified ELISA and their application. In E3S Web of Conferences (Vol. 290, p. 03020). EDP Sciences.
- Zhang, S., Garcia-D’Angeli, A., Brennan, J. P., & Huo, Q. (2014). Predicting detection limits of enzyme-linked immunosorbent assay (ELISA) and bioanalytical techniques in general. Analyst, 139(2), 439-445.
- Bishop, J. D., Hsieh, H. V., Gasperino, D. J., & Weigl, B. H. (2019). Sensitivity enhancement in lateral flow assays: a systems perspective. Lab on a Chip, 19(15), 2486-2499.
- Yanagisawa, N., & Dutta, D. (2012). Enhancement in the sensitivity of microfluidic enzyme-linked immunosorbent assays through analyte preconcentration. Analytical chemistry, 84(16), 7029-7036.
- Fischer, S. K., Joyce, A., Spengler, M., Yang, T. Y., Zhuang, Y., Fjording, M. S., & Mikulskis, A. (2015). Emerging technologies to increase ligand binding assay sensitivity. The AAPS journal, 17, 93-101.
- Zhang, Y., Gu, H., & Xu, H. (2024). Recent progress on digital immunoassay: how to achieve ultrasensitive, multiplex, and clinically accessible detection? Sensors & Diagnostics.
- Liu, C. C., Lin, C. C., Liou, M. H., Hsiao, Y. C., Chu, L. J., Wang, P. J., … & Yu, J. S. (2024). Development of antibody-detection ELISA based on beta-bungarotoxin for evaluation of the neutralization potency of equine plasma against Bungarus multicinctus in Taiwan. International Journal of Biological Macromolecules, 262, 130080.
- Aghamolaei, S., Mamaghani, A. J., Ashrafi, K., Kazemi, B., Bandehpour, M., Rouhani, S., … & Tabaei, S. J. S. (2024). Designing and Developing Serological Test for the Diagnosis of Human Fascioliasis Using a New Recombinant Multi-epitope. Acta parasitologica, 69(1), 1005-1015.
- Steel, R., Botteon, A., & Timms, M. Techniques for improving the specificity of sandwich enzyme‐linked immunosorbent assay‐based drug screening. Drug Testing and Analysis.