Microbial Ecology
Microbial ecology is a field of study that examines the relationship between microorganisms and their surroundings. This includes investigating the interactions between microorganisms and the physical, chemical, and biological factors of their environment, as well as their role in ecological processes.
Microorganisms, such as bacteria, fungi, and viruses, are present in various habitats and are crucial for the proper functioning of ecosystems by contributing to nutrient cycling and energy flow.
The goal of microbial ecology is to comprehend the diversity, distribution, and behavior of microorganisms in different environments, as well as their response to changes in the environment.
Research in this field employs a range of methods such as microbial culturing, molecular biology, metagenomics, and bioinformatics. The study of microbial ecology can provide valuable insights into ecosystem functioning and offer solutions for managing and preserving microbial diversity. The applications of microbial ecology extend to areas like bioremediation, biotechnology, and medicine.
Pollution Monitoring
Pollution monitoring using PCR (polymerase chain reaction) is a technique that uses genetic material to detect and quantify pollution in environmental samples. PCR is a highly sensitive and specific method that amplifies and detects specific DNA sequences, allowing for the detection of even small amounts of target DNA in a sample.
In pollution monitoring, PCR is often used to detect and quantify the presence of specific microorganisms or genes that are indicative of pollution. For example, the presence of fecal indicator bacteria such as E. coli in water samples can indicate contamination by sewage or animal waste.
To perform PCR-based pollution monitoring, environmental samples such as water, soil, or air are collected and processed to extract DNA. The extracted DNA is then amplified using PCR with specific primers that target the genes of interest. The PCR products are then analyzed to determine the presence and abundance of the target DNA.
Overview of PCR and its Applications in Environmental Studies
Purification of nucleic acids from Environmental Microorganisms for PCR Amplification
- To successfully amplify DNA using PCR from environmental microorganisms, it is crucial to extract and purify nucleic acids with care. Environmental samples often contain various organic and inorganic materials that can interfere with the PCR reaction, such as humic materials, clay, and other organics.
- These contaminants can bind to DNA and inhibit the activity of the DNA polymerase, or bind to the DNA template and prevent amplification. As little as 0.001 μg of montmorillonite humic material added to the PCR reaction can cause inhibition. To overcome these inhibitory effects, it is necessary to carefully extract and purify nucleic acids from environmental samples using appropriate methods such as silica-based column purification or magnetic bead-based purification.
- PCR inhibitors such as BSA can be used, and reaction conditions may need to be modified to minimize the effects of environmental contaminants. It is also important to include positive and negative controls in PCR reactions to monitor for contamination or other issues that may affect amplification.
Purification of Nucleic Acids from Soil and Sediment
- Methods for extraction and purification include direct lysis or isolation of bacterial cells, followed by a combination of standard methods such as phenol-chloroform extraction, dialysis, chromatography, and density gradient centrifugation.
- RNA isolation and purification can be achieved using guanidinium hydrochloride and phenol-chloroform-isoamyl alcohol extraction followed by ethanol precipitation.
- Direct lysis can result in higher nucleic acid recovery but can also introduce eukaryotic DNA contamination.
Purification of Nucleic Acids from microorganisms in water
Some of the methods include collecting cells, lysing them, and separating the nucleic acids using filter cartridges, CsCI-EtBr density gradient centrifugation, PVPP treatment, and repeated phenol-chloroform extractions.
PCR methodology for environmental applications
The basic methodology of PCR in environmental applications includes,
- “Hot start,” – The “hot start” method is used in PCR to improve specificity by minimizing the formation of unwanted DNA bands. It works by preventing nonspecific priming and extension at lower temperatures before the reaction reaches the optimal temperature for DNA synthesis.
By activating the DNA polymerase only at higher temperatures, nontarget amplification can be minimized, and primer dimer formation can be reduced. This method can be achieved either by modifying the reaction conditions or by using specialized DNA polymerases that require high temperatures for activation.
- Removing PCR contaminants – More effective measures like treating the reaction mixture with restriction enzymes, DNase I, or modifying contaminating DNA with psoralen or isopsoralen can be taken. The substitution of dUTP for dTTP in PCR reactions and treatment with uracil DNA glycosylase can also eliminate dU-containing DNA, but this approach may not work if RNA is present, and the newly amplified DNA product needs to be stored at a high temperature to prevent enzyme activity.
- Thermostable DNA polymerases from thermotolerance microorganisms with additional activities such as reverse transcriptase – Several types of thermostable DNA polymerases can be used for PCR amplification, including native Taq DNA polymerase and recombinant AmpliTaq DNA polymerase of Thermus aquaticus.
However, newer polymerases such as the Stoffel fragment, Tth polymerase, Vent DNA polymerase, and Pfu DNA polymerase have potential advantages for environmental PCR technology due to their increased thermo stability.
Detection 0f Indigenous Microorganisms In The Environment By PCR
- Detection of Degrading Microorganisms
The study designed oligonucleotide primers based on the nucleotide sequence of a chlorocatechol dioxygenase-degrading gene (tfdC) from Alcaligenes eutrophus JMP134 (pJP4). These primers were used for the detection of different chloro-aromatic-degrading bacteria by PCR amplification.
- Identification of Microorganisms in Biofilms
PCR was used to detect the population architecture of Gram-negative sulfate-reducing bacteria in a sulfidogenic biofilm established in an anaerobic fixed-bed bioreactor. The PCR amplification targeted a conserved region of the 16S ribosomal RNA gene that is present in resident sulfate-reducing bacteria.
- Detection of Indicator Microorganisms in Water
A PCR gene probe-based method for the detection of coliform bacteria and the amplification of a portion of the lacZ gene detects E. coli and other coliform bacteria, including Shigella species.
Case studies
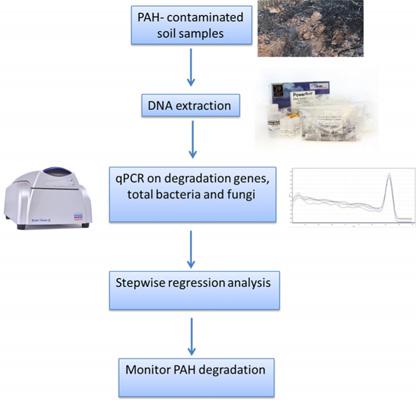
The protocol outlines a method for quantifying functional genes involved in the degradation of PAHs (polycyclic aromatic hydrocarbons) in soil samples using qPCR (quantitative polymerase chain reaction). PAHs are toxic pollutants with carcinogenic and mutagenic properties, and their clean-up is essential. Bioremediation, which involves the use of PAH-degrading microorganisms, is an effective and cost-efficient method for degrading PAHs. These microorganisms contain functional genes that enable them to use PAHs as a source of food and energy.
Traditionally, culture-based methods such as most probable number (MPN) and plate counting have been used to count PAH degraders. However, these methods only capture a small fraction (<1%) of microorganisms capable of PAH degradation. Hence, culture-independent methodologies are preferred.
The qPCR method presented in the protocol is fast, sensitive, and reliable for quantifying the functional genes involved in PAH degradation in soil samples. It is based on the detection of specific DNA sequences using fluorescent probes and allows for the detection and quantification of specific microbial populations without the need for culturing. The method involves extracting DNA from soil samples, amplifying the target gene using qPCR, and quantifying the results using a standard curve. The protocol provides detailed instructions for DNA extraction, primer and probe design, qPCR amplification, and data analysis.
This protocol provides a culture-independent method for the quantification of functional genes involved in PAH degradation in soil samples using qPCR. It offers a fast, reliable, and sensitive approach to monitoring the effectiveness of bioremediation strategies and assessing the impact of pollutants on the environment.
References
- Tsai, Y., M.J. Park, and B.H. Olson. 1991. A rapid method for direct extraction of mRNA from seeded soils. Appl. Environ. Microbiol. 57: 765-768.
- Paul, J.H., L. Cazares, and J. Thurmomd. 1990. Amplification of the rbcL gene from dissolved and particulate DNA from aquatic environments. Appl. Environ. Microbiol. 56: 1963-1966.
- Weller, R. and D.M. Ward. 1989. Selective recovery of 16S rRNA sequences from natural microbial communities in the form of cDNA. Appl. Environ. Microbiol. 55: 1818-1822.
- Mullis, K.B. 1991. The polymerase chain reaction in an anemic mode: How to avoid cold oligodeoxyribonuclear fusion. PCR Methods Applic. 1: 1-4.
- Longo, M.C., M.S. Berninger, and J.L. Hartley. 1990. Use of uracil DNA glycosylase to control carry-over contamination in the polymerase chain reaction. Gene 93: 125-128.
- Greer, C.W., D. Beaumier, H. Bergeron, and P.C.K. Lau, personal communication.
- Amann, R.I., J. Stromley, R. Devereux, R. Keryl, and D.A. Stahl, in preparation.
- Bej, A.K., R.J. Steffan, J.L. DiCesare, L. Haft, and R.M. Atlas. 1990. Detection of coliform bacteria in water by a polymerase chain reaction and gene probes. Environ. Microbiol. 56: 307-314.