Advances in genome sequencing technology have had a significant impact on agriculture and food security. With the ability to sequence the genomes of various crops, livestock, and microbes, researchers can gain valuable insights into the genetic makeup of these organisms, which can help improve their productivity, disease resistance, and nutritional value. Genome sequencing also plays a critical role in ensuring food safety by allowing for the rapid detection of foodborne pathogens and the identification of their sources. In this context, genome sequencing has emerged as a powerful tool to address the challenges of feeding a growing population while minimizing environmental impact and ensuring food safety.
Importance of Genome Sequencing in Agriculture and Food Security
- The global food supply is complex and has become more so due to factors such as increased availability of minimally and highly processed foods and longer distances traveled by food.
- Consumption patterns and increased susceptibility of certain populations have increased the risk of foodborne illnesses, causing concerns about the safety of the food supply.
- Foodborne diarrheal diseases are a significant concern, with more than 2000 children dying every day due to diarrhea caused by contaminated food or water.
- Surveillance and estimation of the burden of foodborne illnesses differ between developing and developed countries, with many cases in developing countries going unreported or unknown.
- Major foodborne pathogens in developed countries include Campylobacter, Salmonella, verocytotoxigenic E. coli (VTEC), and Listeria monocytogenes, with significant economic impact.
- Technologies and programs have been developed and implemented in the food industry to mitigate the risk of foodborne diseases, but some pathogens have shown increased cases in certain regions.
- There is a need for novel mitigation strategies that combine scientific fields such as genomics, modeling, ecology, epidemiology, and statistics to address these challenges.
Genome Sequencing
A reference genome contains comprehensive information on the nucleotide sequence of all chromosomes and provides structural details through annotations. Annotations describe the relative arrangement of genes about non-coding sequences, centromeres, and chromosome ends.
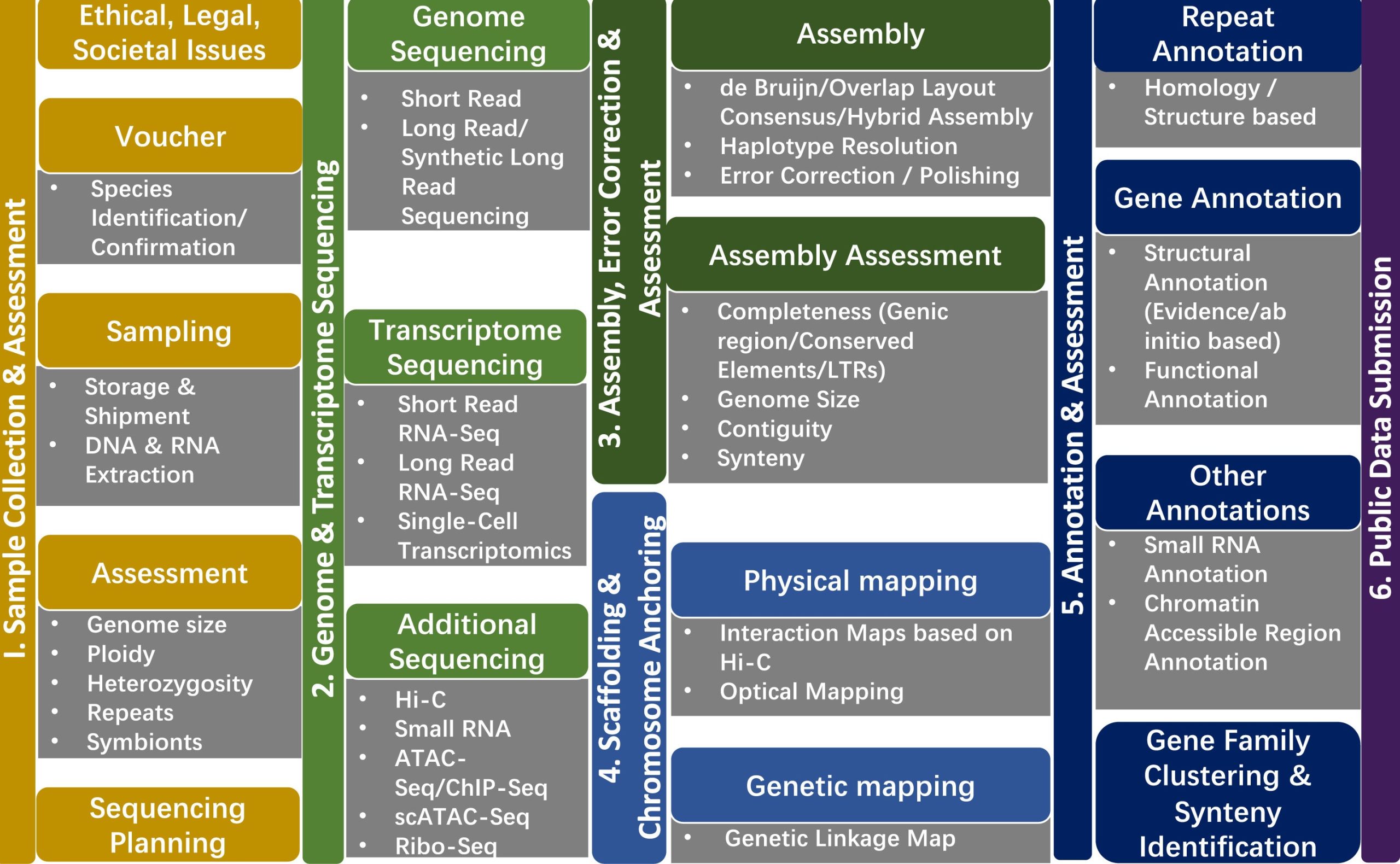 A generalized plant genome workflow from sample collection through assemblyand annotation to public data submission.
A generalized plant genome workflow from sample collection through assemblyand annotation to public data submission.
Applications of Genome Sequencing in Agriculture
- Synthetic biology is a new strategy to develop novel agronomic traits quickly. The CRISPR-Cas system has the potential for improving plant design and synthetic biology.
- Endogenous genes can be edited or foreign genes can be introduced to redirect metabolic networks or establish new pathways in plants. CRISPR-Cas-mediated multiplex gene editing and regulation could be used for synthetic biology.
- Photosynthesis systems in plants are inefficient, but CRISPR-mediated DNA insertion could increase efficiency by introducing components to bypass photorespiration or redesigning Rubisco.
- Genome editing could facilitate other aspects of plant synthetic biology, such as building plant biosensors to monitor intracellular signals or plant recorders to detect environmental stimulation. Research on Arabidopsis thaliana has greatly enhanced the understanding of plant development and stress tolerance, and its genes can be used as candidates for identifying orthologs in crops. However, this approach is not very effective for disease resistance because there are two resistance mechanisms, PTI and ETI, with ETI being specific to each species.
- The draft genome of H. brasiliensis, which is approximately 2.1 Gb in size, was recently published to gain more understanding of its noncoding regions and regulatory roles (Rahman et al. 2013). The assembly comprises 1.1 Gb of scaffolds and 78% of the genome was estimated to be repetitive DNA. There were 68,955 gene models predicted, of which 12.7% are unique to H. brasiliensis. Most of the genes involved in rubber biosynthesis, rubber wood formation, disease resistance, and allergenicity have been identified using this genomic information, along with transcriptomic studies.
Breeding and Crop Improvement
- Tropical countries often have lower levels of development compared to temperate countries, largely due to poor agricultural productivity. Many important crops originated in the tropics, but colonial rule and a lack of attention to staple food crops have reduced the variety of crops grown.
- The green revolution of the late 1960s used an integrated approach to improve crop productivity through the use of improved varieties, fertilizers, and pesticides. Hybridization and transgenic technology have also been used to create high-yielding crop varieties, although the latter is controversial due to concerns about potential health and environmental risks Guimaraes E P. (2009).
- Next-generation sequencing technologies have revolutionized the field of genetics and offer new opportunities for crop breeding and improvement. In this article, we review the applications and limitations of these technologies and discuss their potential for future research in crop improvement. We also highlight the important crop genomes that have been sequenced to date.
- Next-generation sequencing technologies have advanced genome sequencing and marker development.
- SNPs are now the dominant molecular marker application, with in silico methods allowing for cheaper and more efficient SNP discovery.
- These markers have been used for developing molecular genetic and physical maps and identifying genes or quantitative trait loci controlling important traits.
- High-density genetic maps are now possible due to advancements in NGS, allowing for the prediction of the linear arrangement of markers in a chromosome.
- Association mapping is now replacing traditional quantitative trait locus mapping due to the high resolution it provides for marking traits.
Genome Sequencing and Livestock Breeding
- The genetic diversity of native cattle breeds from the northernmost region of cattle farming was investigated through whole-genome sequencing for the first time.
- Novel SNPs and indels and unannotated genes were discovered, highlighting the need for accurate reference genome assemblies for genetically diverse native cattle breeds.
- Identification of genes and chromosomal regions important for adaptation and production traits was made.
- Enriched GO terms in genes associated with selective sweeps include defense response, growth, sensory perception, and immune response.
- More animals of these breeds should be investigated in a wider breed diversity context to enhance knowledge of their genetic resources for future cattle breeding and the power of selection signature analyses.
Pathogen Detection and Identification
The use of whole-genome sequencing (WGS) has become widespread for bacterial sequencing. The complete genome of a prokaryote offers information about its potential phenotypes. Therefore, there is no need for a preliminary classification based on taxonomy, followed by an inference of the likelihood of the pathogen’s virulence. If the genome sequence can be established, along with the presence of virulence and pathogenicity factors, then WGS can provide an extremely high-resolution assay, which can differentiate between two isolates at the single-base level. As a result, it is an excellent tool for precise tracking of outbreaks and transmission, providing unparalleled opportunities.
References
- ” Food safety trends: From the globalization of whole genome sequencing to the application of new tools to prevent foodborne diseases”, Trends in Food Science & Technology, Volume 57, Part A, November 2016, Pages 188-198.
- Ballouz, A. Dobin, J. A. Gillis, “Is it time to change the reference genome? Genome “Biol. 20, 159 (2019).
- Chen, Y. Wang, R. Zhang, H. Zhang, C. Gao, “CRISPR/Cas Genome Editing and Precision Plant Breeding in Agriculture “, Annu. Rev. Plant Biol., 70 (2019), pp. 667-697
- F. South, A.P. Cavanagh, H.W. Liu, D.R., “ Ort Synthetic glycolate metabolism pathways stimulate crop growth and productivity in the field Science”, 363 (2019), p. eaat9077.
- H. Gunn, E. Martin Avila, R. Birch, S.M. Whitney, “ The dependency of red Rubisco on its cognate activase for enhancing plant photosynthesis and growth”, Proc. Natl. Acad. Sci. USA, 117 (2020), pp. 25890-25896.
- Feuillet C, Leach J E, Rogers J, Schnable P S and Eversole K. (2011), “Crop genome sequencing: Lessons and rationales”, Trends in Plant Science 16(2): 77–88.
- Rahman A Y A, Usharraj A O, Misra B B, Thottathil G P, Jayasekaran K, Feng Y, Hou S, et al. (2013), “Draft genome sequence of the rubber tree Hevea brasiliensis”, BMC Genomics 14(1): 75.
- M J Carena (ed.), “Cereals, the banks, and the Italian economy”, USA: Springer Science+Business Media LLC, 99–126.
- Pérez-de-Castro A M, Vilanova S, Cañizares J, Pascual L, Blanca J M, Díez M J, ProhensJ and Picó B. (2012).” Application of genomic tools in plant breeding. Current Genomics”, 13(3): 179–195.
- Melak Weldenegodguad, “Whole-Genome Sequencing of Three Native Cattle Breeds Originating From the Northernmost Cattle Farming Regions”, Front. Genet., 11 January 2019 Sec. Livestock Genomics, Volume 9 – 2018 |