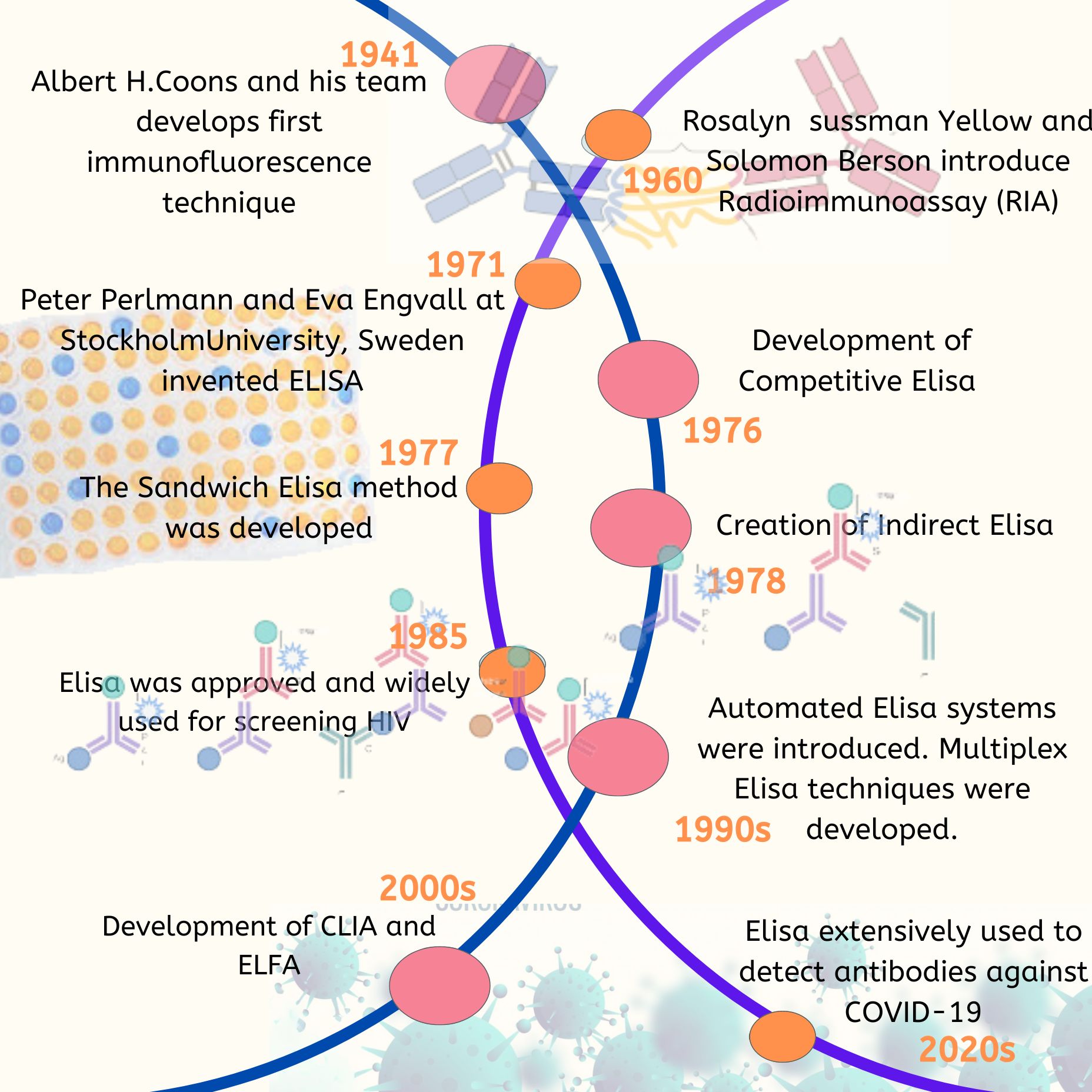
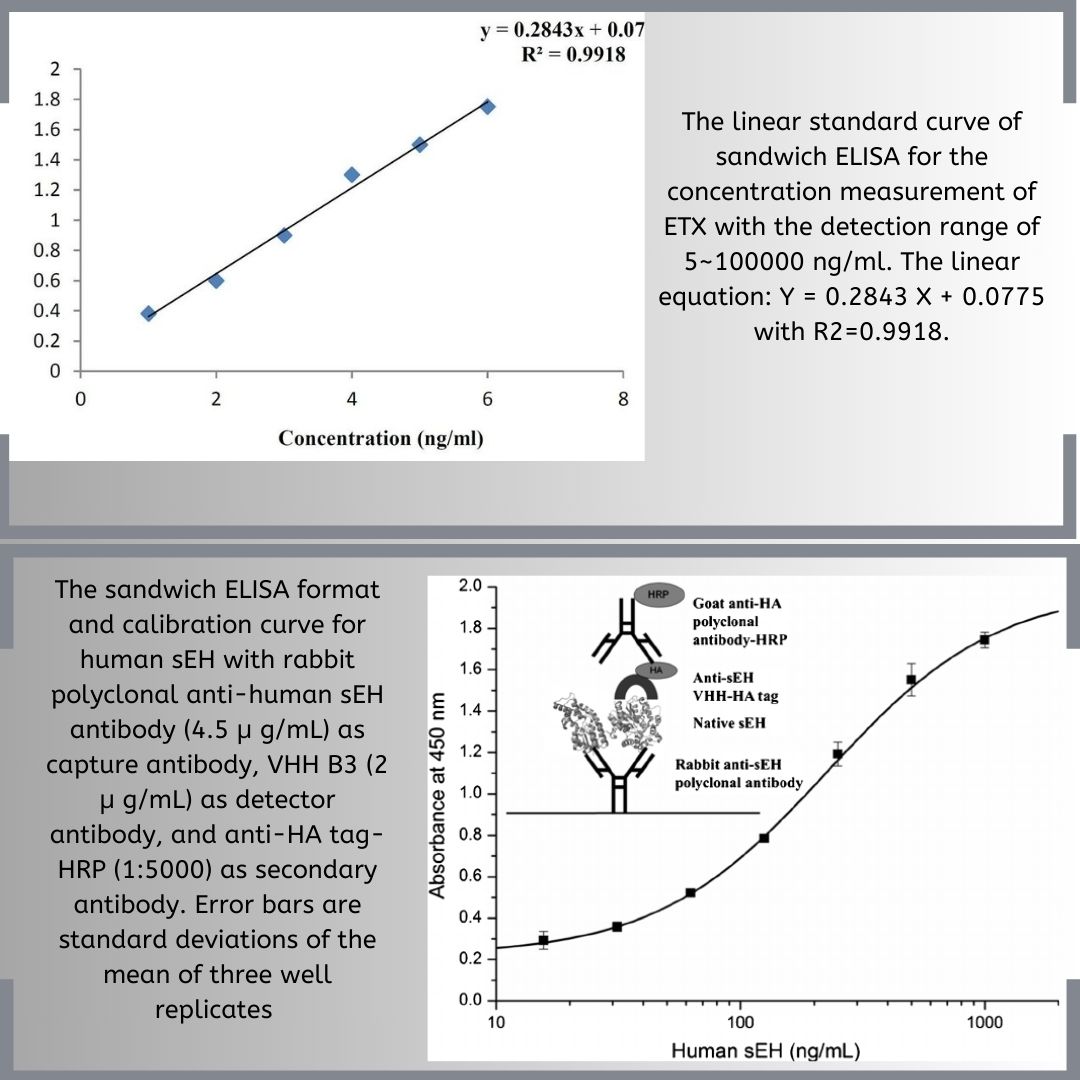
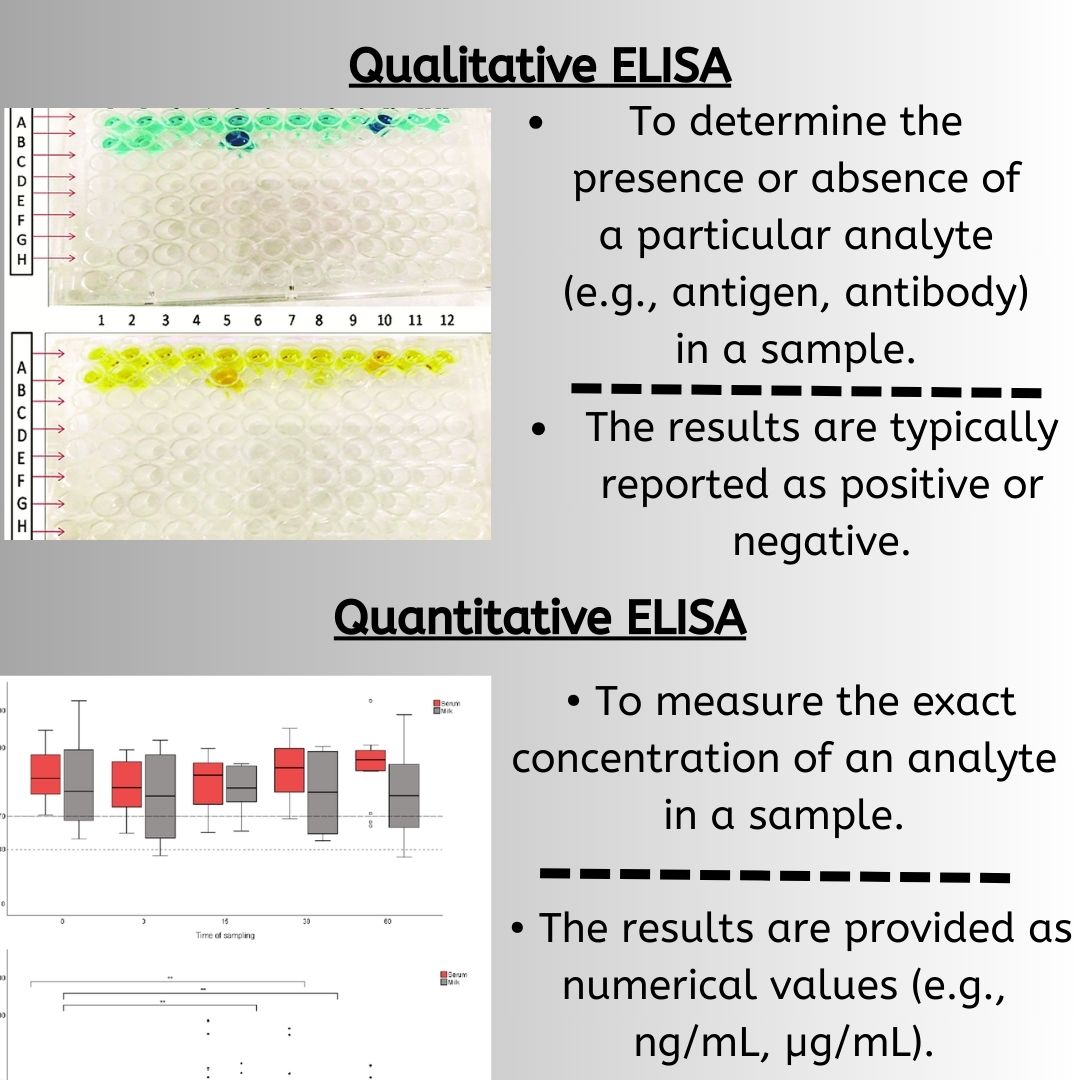
The process of Polymerase Chain Reaction (PCR) is an innovative approach that allows for the amplification of samples to aid in their identification and analysis. In the fields of life sciences and medicine, PCR has grown to become an influential tool. Its applications include the detection of pathogens, either specific or broad-spectrum, monitoring emerging infections, analyzing antimicrobial resistance, screening for neonatal genetic disorders, and identifying genes linked to tumors.
PCR has had a remarkable impact on our comprehension of biology and medicine, as it provides quick and accurate results that are dependable. One of the primary advantages of PCR is its speed, as it can quickly amplify target samples from a single source through the exponential growth of DNA fragments in the reaction. This can reduce the time taken to obtain results from days or weeks to just a few hours. Furthermore, PCR assays are highly sensitive, making it possible to identify and analyze small samples such as single cells without requiring a vast amount of the specimen material.

PCR has another benefit in that it provides excellent precision, particularly with techniques such as Real-Time Quantitative PCR, which can yield significant data on transcript levels or gene expression. With various assay options at its disposal and the ability to design primers targeting specific genes, PCR can achieve highly specific detection of target cases, even in complex interactions that involve multiple genes from different species.
Although PCR is a powerful technique, sometimes PCR reactions can fail due to various reasons. Here are some common PCR troubleshooting problems and their solutions:
-
No Amplification or Low Yield:
No amplification or low yield in PCR is a common problem that researchers encounter during PCR experiments. The first step in troubleshooting no amplification or low yield in PCR is to confirm the presence of the DNA template. This can be done by measuring the concentration and purity of the DNA template using spectrophotometry or fluorometry. If the DNA concentration is low or the purity is poor, it can affect the success of the PCR reaction. In this case, the DNA template may need to be purified or concentrated before proceeding with the PCR reaction.
The next step is to optimize the PCR conditions. This involves adjusting the annealing temperature, the MgCl₂ concentration, and the reaction buffer. If the annealing temperature is too low or too high, it can result in non-specific amplification or no amplification, respectively. The MgCl₂ concentration affects the activity of the polymerase enzyme and can affect the efficiency of the PCR reaction. The reaction buffer contains salts and other components that are necessary for the PCR reaction to proceed. If the buffer is suboptimal, it can affect the success of the PCR reaction.
Also, the amount of enzyme and dNTPs can affect the success of the PCR reaction. If the amount of enzyme or dNTPs is too low, it can result in no amplification or low yield. Increasing the amount of enzyme or dNTPs can improve the efficiency of the PCR reaction.
-
Non-Specific Products
Non-specific amplification occurs when the primers bind to unintended regions of the template DNA, resulting in the amplification of nonspecific products.
Some polymerases exhibit low activity when exposed to room temperature or 4 °C. During the mixing of different reaction components, primers may anneal non-specifically, leading to the enzyme elongating these primers and generating a series of non-specific products. To prevent these non-specific products, hot-start polymerases can be employed. Hot-start enzymes can be created through various techniques, such as manual or physical separation, antibodies, or chemical modification. In the manual technique, one of the reaction mixture components, such as Mg2+, is added to the tube after the temperature exceeds 70 °C. Physical separation involves the use of a barrier, such as a wax plug, to separate reaction components until the temperature rises above 75 °C, at which point the wax melts and the components are mixed. Another method involves inactivating polymerases through heat-sensitive antibodies or heat-labile blocking groups that are added to specific amino acids. At higher temperatures, the blocking groups are removed, and the enzyme is activated.
-
Primer-Dimer Formation:
Primer-dimer formation can occur when the primers anneal to each other due to the complementarity between their sequences, resulting in a self-priming event. These self-priming events can be promoted by high primer concentrations, high annealing temperatures, and long annealing times. Primer-dimer formation is a problem in PCR because it can reduce the yield of the target DNA sequence, leading to false negative results, and also can create additional, unwanted products that can interfere with downstream analysis.
There are several ways to prevent the primer-dimer formation in PCR. The first is to optimize the PCR conditions, such as annealing temperature, primer concentration, and annealing time. Increasing the annealing temperature can reduce the likelihood of primer-dimer formation, but it can also decrease the efficiency of primer annealing to the target DNA.
Another approach is to design the primers carefully. Primers should be designed with specificity and a minimum amount of complementarity between themselves. Primers should also be checked for secondary structures, which can promote self-priming events. Software programs are available to help design primers with specificity and minimize the potential for self-priming.
-
Inhibition of PCR:
The inhibitors of the PCR reaction refer to diverse organic and inorganic compounds that may obstruct DNA polymerase directly (by causing polymerase degradation or obstructing the polymerase’s active center) or indirectly (by blocking the active center for cofactors such as magnesium ions) and/or interact with the nucleic acid template.
To minimize the effects of inhibition, several strategies can be employed. The first step is to optimize the PCR conditions, including the concentrations of the reaction components and the cycling conditions. The concentrations of the reactants, such as the template DNA, primers, and DNA polymerase, should be optimized to maximize the efficiency of amplification while minimizing the effects of inhibition.
Another approach is to use specialized PCR additives, such as bovine serum albumin (BSA), to help overcome the effects of inhibition. BSA can help reduce the binding of inhibitors to the DNA polymerase, thereby improving its activity. Other additives, such as betaine, can also be used to reduce the effects of inhibition by destabilizing the secondary structure of the template DNA.
-
Uneven or Smeared bands:
One of the primary causes of uneven or smeared bands in PCR is suboptimal PCR conditions. If the annealing temperature is too low, non-specific products can be generated, leading to smeared bands on the gel. If the extension time is too long, secondary products can be formed, which can contribute to smearing.The quality of the template DNA can also affect the appearance of the PCR product on the gel. Degraded DNA can lead to the formation of shorter and larger fragments, which can contribute to smearing. Contaminants in the DNA sample can also interfere with PCR amplification, leading to smearing.
According to a study, the issue of PCR smears during genotyping is caused by the gradual accumulation of “amplifiable DNA contaminants” that are specific to the PCR primers being used. As a result, previously reliable primers are no longer effective once smears appear. To address this problem, it is recommended to take preventive measures such as separating lab areas, reagents, and equipment for pre-PCR from post-PCR to slow down contamination buildup. However, the most efficient solution is to switch to a new set of primers with different sequences that do not interact with the accumulated contaminants, thus completely resolving the smearing issue.

Important Considerations When Troubleshooting PCR
- If the standard conditions for PCR do not produce the intended amplicon, it is necessary to optimize the PCR process for better outcomes. The specificity of the reaction can be adjusted by modifying variables such as reagent concentrations and cycling conditions to achieve the desired amplicon profile. For instance, if the reaction is not stringent enough, multiple spurious amplicons of varying lengths may be produced, whereas a reaction that is too stringent may not produce any product at all. Although troubleshooting PCR reactions can be frustrating, careful analysis and a comprehensive understanding of the reagents used in the experiment can shorten the time and number of trials required to obtain the intended results.
- Among the factors that affect PCR stringency, adjusting the concentration of Mg2+ and/or modifying annealing temperatures are likely to address most issues. Nevertheless, it is important to ensure that any erroneous result was not caused by human error before making any changes. It is recommended to first verify that all necessary reagents were added to the reaction and that they were free from contamination.
- It is crucial to identify if any of the PCR reagents are harmful to the reaction. To do so, prepare fresh working stocks or dilutions of the reagents and add them one at a time to the reaction mixture systematically. This procedure will help to pinpoint which specific reagent caused the unsuccessful PCR experiment.
In general, PCR has several inherent benefits over other techniques for observing minute changes at both macro and micro levels in cells and tissues that were previously difficult or costly to detect. However, any application involving PCR must consider its pros and cons before attempting its use, as neglecting to do so could result in suboptimal outcomes due to its intricate nature.
References
- Polymerase Chain Reaction. Molecular Biology, 168–198 | 10.1016/B978-0-12-813288-3.00006-9
- Śpibida, M., Krawczyk, B., Olszewski, M., & Kur, J. (2016). Modified DNA polymerases for PCR troubleshooting. Journal of Applied Genetics, 58(1), 133–142. doi:10.1007/s13353-016-0371-4
- https://www.jove.com/t/3998/polymerase-chain-reaction-basic-protocol-plus-troubleshooting
- Cause and solutions to the polymerase chain reaction smear problem in genotyping Dawn D. Han a , Rong Chen a , Erik R. Hill a , Michael R. Tilley a , Howard H. Gu a,b,¤ a Department of Pharmacology, The Ohio State University College of Medicine, 333 West 10th Avenue, Columbus, OH 43210, USA
- Wang M, Cai J, Chen J, Liu J, Geng X, Yu X, et al. PCR Techniques and Their Clinical Applications [Internet]. Polymerase Chain Reaction [Working Title]. IntechOpen; 2023. Available from: http://dx.doi.org/10.5772/intechopen.110220